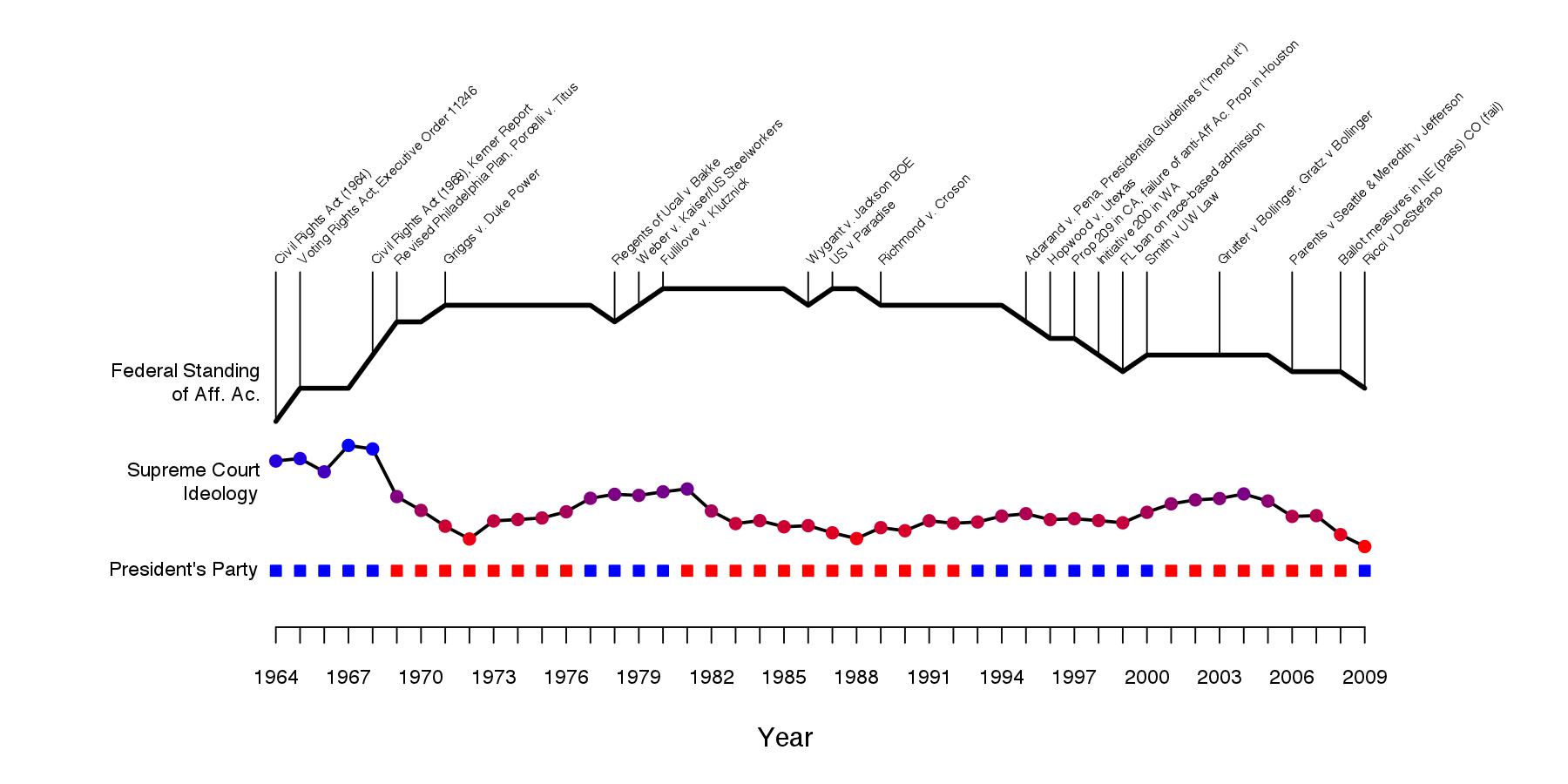
An heuristic visualization for use in my class on comparative political economy of affirmative action policies. Federal standing is scored simply by adding a point for each federal action or Supreme Court decision promoting the application of affirmative action, and subtracting a point for each action that curtails it. It is a gross simplification of course, but by appearances, it seems to capture the trends well. For example, while there have been steady curtailments since the early 1990s, it would be inaccurate to suggest federal standing is where it was at the time of enacting the Civil Rights Act in 1964. The federal level actions causing the score to increase or decrease are also shown.
Below the federal standing score is a plot of the Martin-Quinn score (link) of the median Supreme Court justice, a measure of the court’s liberal orientation (shown as blue, higher values) versus conservative orientation (red, lower values). Below that is a timeline of the party of the president.
John David Skrentny, in his magisterial account of the evolution of affirmative action politics in the US (link), highlights crisis management in the face of the race riots of the mid-1960s and US leadership’s concerns over international reputation during the Cold War as crucial determinants of affirmative action’s rise in that period. We can see that, at least by measure of party and court ideology, the institutional/ideological context was also especially receptive. At the same time, trends pushing upward the federal standing of affirmative action continued despite Nixon assuming office and a trend toward conservatism on the court.